Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Jannatul Ferdous and Hamida Khanum*
Received: November 14, 2022; Published: November 29, 2022
*Corresponding author: Hamida Khanum, Department of Zoology, University of Dhaka, Dhaka 1000, Bangladesh
DOI: 10.26717/BJSTR.2022.47.007487
Though the bacterium inhabited in lower intestinal tract often discharged into environment through faeces or wastewater effluent. The presence of E. coli in surface water is a sign of feacal contamination. Main focus of this study was to isolation and identification of pathogenic strains of E. coli in the pagla sewerage water treatment plant of Dhaka city, to understand the effectiveness of the sewerage treatment plant. Water samples were collected and total 24 number collected water samples 480 E. coli strains were isolated. Various cultural and biochemical tests were performed to isolate E. coli. An important aspect of this study was to examine the use of specific fluorescent dye 4-methyl-umbelliferone (MUG). 40 E. coli isolates were selected for MUG test among them 37 isolates were positive, which means 92.50% E. coli isolates were MUG test positive. Another important feature of this study was to examine E. coli isolates in CR-SMAC (Sorbitol- MacConkey agar containing cefixime and rhamonose) plate. CR-SMAC is the world recognized selective medium for E. coli 0157:H7. In SMAC, lactose has been replaced by sorbitol. E. coli 0157:H7 do not ferment sorbitol and produce colorless colony; on the other hand most other E. coli ferment sorbitol and thus produces pink colony. Among 480 E. coli isolates 80 E. coli isolates were isolated from raw water point, were selected and after screening was done by plate technique and after biochemical test 40 E. coli strains were selected for multiplex PCR. The results of multiplex PCR showed that, two pathogenic strains (376 bp and 322 bp) were present in the raw water point. Data presented that as virulent marker genes were absent at the final water point so the treatment plant works effectively.
Keywords: Sewage Treatment Plant; Pathogenic Strain; Surface Water; Escherichia Coli 0157:H7
Abbreviations: MUG: Methyl-Umbelliferone; CR-SMAC: Sorbitol- Macconkey Agar Containing Cefixime And Rhamonose; ETEC: Enterotoxigenic E. coli; WHO: World Health Organization; EPEC: Enteropathogenic E. coli; EIEC: Enteroinvasive E. coli; EAggEC: Enteroaggregative Escherichia coli; CDEC: Cell Detaching E. coli; EHEC: Enterohaemorrhagic Escherichia Coli; PCR: Polymerase Chain Reaction; PET: Polyethylene Terephthalate; PCR: Polymerase Chain Reaction; EML: Environmental Microbiology Laboratory; TC: Total Colifrom; FC: Faecal Colifrom; FS: Faecal Streptococci; MFC: Membrane Faecal Coliform; CFU: Colony Forming Unit; LT: Heat-Labile; HUS: Haemorrhagic Urenic Syndrom; UTI: Urinary Tract Infection
E. coli is a rod shaped, Gram-negative bacterium in the Family Enterobacteriaceae is the commonly inhabits the lower intestinal tract of humans and animals as well as one of the significant pathogen (Lorenz, et al. [1-3]). The organism usually inhabits the infant gastrointestinal tract within hours of birth and afterward E. coli and the host descend mutual assistance (Nataro, et al. [4,5]). Though E. coli usually confined to the intestinal lumen but in case of immune suppressed patients can be infected by nonpathogenic strains of E. coli. Most E. col istrains live offensively in the human intestine as intestinal flora. However pathogenic strains can cause severe diarrheal diseases in healthy individuals. Pathogenic E. coli generally caused (Allocati, et al. [3,6]).
i) Urinary tract infection
ii) Meningitis and
iii) Enteric or diarrheal disease
Diarrheal diseases are one of the major public health problems due to high rate of morbidity in infants in our country; however, it is the predominant reasons of mortality, ensuing in about two million deaths worldwide each year (Alikhani, et al. [7]). World Health Organization (WHO) estimated that 80% diseases are caused by inadequate sanitation, contaminated water or unavailability of safe water in our country. E. coli strains can survive on these sources executes direct relationship to the incidence of the disease.Highly adaptive E. coli clones have the capacity to cause an extensive range of diseases. Survival is related with maintenance of viability under unfavorable environment (Pickup [8]). When E. coli is released to the environment it encounters nutritional and physicochemical shocks or stresses affecting short duration of survival. (Savageau, et al. [9-11]). Recent research discovers that E. coli can survive in the environment, soil, water for almost a month’s (Devane, et al. [12]). Not only E. coli , other species like Klebsiella spp, Vivrio spp and Candida albicans also able to survive in tropical waters and may multiply (Lopez- Torres, et al. [13,14]).
According to distinct epidemiological and clinical features five specific virulence serotypes identified which are Enterotoxigenic E. coli (ETEC), Enteropathogenic E. coli (EPEC), Enteroinvasive E. coli (EIEC), Enterohaemorrhagic Escherichia coli (EHEC), Enteroaggregative Escherichia coli (EAggEC) (Nguyen, et al. [15,3]). Other diarrhoeagenic E. coli pathotypes have been proposed, for example diffusely adherent E. coli (DAEC), Cell Detaching E. coli (CDEC) (Huang, et al. [16]). Different diagnostic methods are developed for identify E. coli such as culture-based enumeration technique and molecular methods remain the reliable techniques like Polymerase Chain Reaction (PCR). E. coli identification in water samples previously described (Martins, et al. [17]). So, the present study was carried out to sort out the genetic occurrence of virulence-producing factors E. coli isolates from water sample by discovering the presence of virulence genes by PCR.
Identification of Virulence Prospective of E. coli
E. coli is a significant indicator of entero-pathogenic contamination and well-studied bacterium as standardized data exist. E. coli is an organism whose presence is thought to be related with pollution and presence of pathogens. But their abundance and genetic diversity in association yet to be discovered, so the focus of this study was lead with the aim of isolating, studying abundance of E. coli and their genetic diversity in sewage water treatment plant using classical microbiology and molecular biology technique. Approach was made to identify five major pathogenic strains of E. coli by multiplex PCR.
Samples were collected from sewage water and sample sites are selected at four different point of pagla sewagetreatment plant. Samples were collected aseptically in sterile polyethylene terephthalate (PET) bottles. They were conveyed to the laboratory as fast as possible and were preserved in appropriate temperature. Soon after arrival samples were analyzed.All the studies were carried out in the Environmental Microbiology Laboratory (EML) under Laboratory Sciences DivEnteroaggregative E. Source of reference strain: Escherichia coli 0157: H7, Enterotoxigenic E. coli (ETEC), Enteropathogenic E. coli (EPEC), Enteroinvesive E. coli (EIEC) and Enteroaggregative E. coli EAEC) were used in this study as positive control which were collected from lab stock.
Reconstitution and Confirmation of the Strain
The strain was streaked on NA plate and incubated at 370C for 24 Hours. Gram staining, 2/3 biochemical test according to the standard procedures reconfirmed the strain. The organism were then stocked and preserved in 30% glycerol broth and kept at – 700C for further study.
Processing of the Sample
Before making any dilution, each of the water samples wasshaken vigorously. 100μl of water sample was added to 900μl of sterile normal saline to give a 10-1 dilution. Then 100μl of the sample from 10-1 dilution was transferred to 900μl of sterile normal saline to prepare a 10-2 dilution. In this way, ten –fold serial dilution of water sample was made from 10-1 to1 to 10-5 by using sterile normal saline. Each dilution was thoroughly mixed using vortex mixture.
Short-Term Preservation
The pure culture was streaked on NA plates and grow overnight at 370C. Form the heavy growth area, cells were accumulated scratching with a sterile needle and stabbed in T1 N1 soft Agar in a cryotube or eppendorf tube. Then the sampleswere stocked up at room temperature for short time (within 1 year).
Molecular Techniques Used for Identification of E. coli
Polymerase Chain Reaction (PCR): Polymerase Chain Reaction (PCR) is a vitro method for synthesis of nucleic acid in which a particular segment of DNA can be specifically amplified. Primers hybridize with complementary strands of the target sequence and are oriented so that DNA synthesized by the polymerase proceeds across the region between theprimers. Since the extension products themselves are also complementary and capable of binding primers, consecutive cycles of amplification essentially double the amount of the target DNA synthesized in the previous cycles. The result is an exponential accumulation of specific target DNA fragments approximately designated by 2n where n is the number of cycles of amplification performed.
Enumeration of Total Colifrom (TC), Faecal Colifrom (FC), Faecal Streptococci (FS) from water sample: Total Coliforms are E. coli, Klebsiella, Citrobacter and Enterobacter. Coliforms are gram negative, rod shaped, non-spore forming bacteria capable of growth in the presence of bile salt or surface active agents with similar growth inhibiting properties, which are cytocrome –oxidase negative and able to ferment lactose at either 350C or 370C with the production of acid, gas and aldehyde within 24 to 48 hours. Membrane faecal coliform (mFC) agar plate was used for the enumeration of coliforms. Then incubated at 370C for 18 to 24 hours. Blue colonies were considered as total coliform (Rashid [18]). Finally, Colony Forming Unit (CFU) was calculated.For the enumeration of fecal coliform same procedure was followed that described for the enumeration of total coliform except the incubation temperature .In case of feacal coliform plates were incubated at 44.50C for overnight and enumerated accordingly. The 0.1ml of samples was spreaded on MacConkey agar plate and incubated at 37⁰C for 24 hours. After 24 hours incubation pink or red color were enumerated as E. coli.
Species-Specific PCR of E.Coli
For species specific detection of E. coli a recently developed primer set is used. The target gene was 60-Mda plasmid. PCR amplification of the target DNA was carried out in a thermal cycler (BIO RAD) using 0.5 ml PCR tubes with a reaction mixture volume of 25 μl. Each of the reaction mixtures contained 1μl template DNA (lysate), 10 pmol of each primer, lμl of 10 pmol deoxynucleoside triphosphates (dNTPs, Invirtogen, USA), 0.25 μl of (5U/μl, Germany) of Taq DNA polymerase, 2.5 μl of 10x PCR reaction buffer (Invitrogen, USA), 0.75 μl of MgCl₂ (50mM, Invitrogen, USA) and nuclease free water to a final volume of 25 μl. The reaction mixture was subjected to an amplification of 35 cycles, according to the following PCR process specification mentioned in the (Tables 1 & 2).
Agarose Gel Electrophoresis of PCR Product
Electrophoresis was carried out in a horizontal gel apparatus. The method followed for agarose gel electrophoresis was described by (Maniatis, et al. [19]). Electrophoresis was conducted in horizontal agarose gels containing 1.0 (Fisons). Agarose (1.0%) was dissolved in TBE (Tris–Borate EDTA) electrophoresis buffer (1x TBE, appendix) by boiling in a micro oven for 5 minutes. Then it was allowed to cool to 55 ⁰C-60 ⁰C and poured into a plastic chamber. Wells were prepared in the gel with combs containing either 8 or 16 teeth. Gel was allowed to solidity for half an hour at room temperature and placed in the electrophoresis tank PCR product (15μl) and tracking dye (3 μl) were mixed with the help of a micropipette and loaded into the wells accordingly. The gel was covered with sufficient amount of 1x TBE buffer before electrophoresis. The power packsupplied regular current. Electrophoresis was carried out at 55 volts until the dye front moved to the end of the gel.
Staining of PCR Product
The gel was stained after run by soaking the gel in 30 μl of ethidium bromide in 150ml of TAE buffer, for half an hour. The gel was destined by washing with 100ml distilled water and DNA bands were viewed on an Ultraviolet Trans illuminator. Gels were photographed by using a Gel Documentation unit. A multiplex PCR for virulence gene of E. coli: A multiplex PCR was performed with 6 sets of primer of which 3 sets identify the presence of virulence factor coding genes present in the core region. Another one is coding for an outer membrane protein winch might have some role in adhesion of in human gut. List of primers and their amplicon size is mentioned in the table. Chromosomal DNA extracted by the above mentioned procedure was used as template for doing PCR in a thermal cycler. Multiplex PCR was performed using 0.2 ml PcR tubes with a reaction mixture volume of 25 μl. Each of the reaction mixture contained 1 μl template DNA (lysate), 10 pmol of each primer, l μl of 10 Mm Deoxynucleoside Triphosphates (dNTPs, Invirtogen, USA) 0.25 μl (5U/μl, Invitrogen, USA) of Taq DNA polymerase, 2.5 μl of 10X PCR reaction buffer (Invitrogen, USA), 0.75 μl of MgC1₂ (50mM, Invitrogen, USA) and nuclease free water to a final volume of 25 μl. The thermal cycler was programmed as follows: initial denaturation a 95⁰C for 5 min, followed by 30 cycles consisting of 95⁰C for 1 min, 55⁰C for 1 min ad 72⁰C for 1 min and a final extension step of 72⁰C for 10 min (Table 3).
Post-Amplification Detection
Agarose- gel-electrophoresis:The post- amplification detection of PCR is performed by agarose- gel- electrophoresis. The successful amplification of virulent gene is examined by resolving the PCR product in 1.2 % agarose gel. After migration for 4h at 60 volts, the gel bromide and. Photographed in an UV- transilluminator). During migration, a 100bp molecular weight marker is always run along with the test sample for measuring the size of DNA band (s) in PCR product. Following migration, DNA band of above mentioned amplicon size was considered as successful amplification of the specific gene.
Abundance of E. coli in raw, inlet, middle and outlet water: The E. coli count was in the range of 4-5.32 log10 cfu/ml in the 24 different studied raw water point while in the inlet water point it was log10 cfu/ml ranged from 3.30-4.54 log10 cfu/ml. The variation of E. coli count was found in the range of 1.54-3.31 log10 cfu/ml in the middle water point and the range was 1.47-2.47 log10 cfu/ ml in the outlet water point. The highest count of E. coli was recorded 5.32 log 10 cfu/ml in the month of May in raw water and the lowest value was recorded in the month of December and the value was 1.47 log10 cfu/ml in outlet water. (Tables 4 & 5). In this study, 24 number water samples were collected and from collected water samples 480 E. coli strains were isolated. An important aspect of this study was to examine the use of specific fluorescent dye 4-methyl-umbelliferone (MUG) that has been available recently for detection of E. coli. E. coli isolates possessing the specific enzyme β-glucuronidase produced flurescence with MUG under long wave length (365nm) of UV light were selected and differentiated from other microorganisms. In this study, 40 E. coli isolates were selected for MUG test among them 37 isolates were positive, which means 92.50% E. coli isolates were MUG test positive. The other important aspect of this study was to examine E. coli isolates in CR-SMAC (Sorbitol- MacConkey agar containing cefixime and rhamonose) plate. CR-SMAC is the world recognized selective medium for E. coli 0157:H7. In SMAC, lactose has been replaced by sorbitol. E. coli 0157:H7 do not ferment sorbitol and produce colorless colony; on the other hand, most other E. coli ferment sorbitol and thus produces pink colony (Table 6).
Result of Multiplex PCR
Determination of virulence genes by multiplex PCR: Template DNA from each strain was subjected to amplification by PCR to detect the presence of eaeA, bfpA and eltB, estA, bfpA, ial and pCVD genes using specific primers (Appendix). Samples that having the genes were given bands of expected size. At first 40 E. coli isolates from raw water point were subjected to a multiplex PCR targeting genes of E. coli core region namely eaeA, bfpA and eltB, estA. Sample no 26 was positive for eaeA gene. Sample 56 was positive for eltB and estAgene.
Detection of eaeA gene by multiplex PCR: Representative E. coli isolates were subjected to multiplex PCR for the detection of specific E.coli virulence genes. eaeA positive isolate was 26 number samples. Positive sample gave 376 bp bands for primer specific to eaeA gene. The result has been shown in the (Figure 1). Detection of eltB and estA gene by multiplex PCR: Representative E. coli isolates were subjected to multiplex PCR for the detection of specific E. coli virulence genes. eltB and estA positive isolate was 56 number sample. Positive sample gave 322 bp bands for primer specific to eltB and estA gene. The result has been shown in the (Figure 2). Next 40 E. coli isolates from outlet water point were subjected to a multiplex PCR including 5 target genes of E. coli core region namely eaeA, bfpA and eltB, estA, bfpA, ial and pCVD. All samples were negative for eaeA, bfpA and eltB, estA, bfpA, ial and pCVD gene. The result has been shown in the (Figure 3). The prevalence of E. coli is the highest in raw water followed by inlet water. The prevalence is negligible in middle water and outlet water compared to raw and inlet water (Table 7).
Figure 2 Note: Lane 1 and 18- Molecular weight marker. Lane2- Positive control (eaeA) Lane4-Positive isolates (26) Lane10- Positive isolates (56) Lane 3 and 11- Negative control Figure 2: Agarose gel Electrophoresis of multiplex PCR
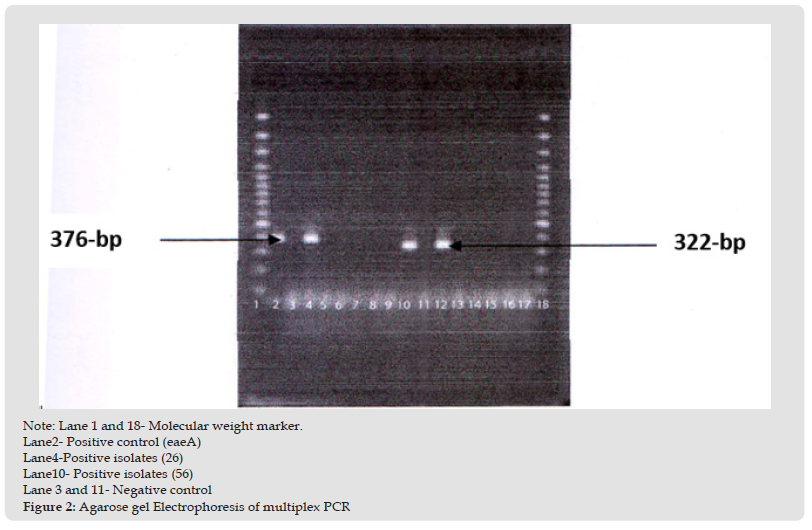
Figure 3 Agarose gel electrophoresis of PCR product. Lane 2, 24 and 25 contain +ve control, Lane 3, -ve controlLane 1 and 18- Molecular weight marker. Lane 3-23 shows the sample And 1, 26 shows molecular Weight marker.
Interpretation
P-value= 0.000 is less than 0.01. This indicates that Anova F-test rejects the null hypothesis of equal revalence of E. coli in different type of water at 1% level of significance. So the prevalence of E. coli may not be equal in Raw, Inlet, Middle and Outlet water. Now it is required to conduct a multiple comparison to test which pair of water type differ significantly in the prevalence of E. coli. Prevalence of E. coli significantly differs in Raw-Middle and Raw-Outlet type water at 1% significance level and all other pairs at 5% significance level except Middle-Outlet pair (Figure 4).
Figure 4 Note: Lane 5-8, 15, 18, 24, 26, shows +control, Lane 1-4 and 9-23 shows the sample Lane 14 and 29 shows molecular weight marker. Figure 4: Agarose gel electrophoresis of PCR product.

Multiple Comparisons by Least Significant Different (LSD) LSD: (Table 8).
Diarrhoeal disease is a foremost cause throughout the world and is accountable for high morbidity and mortality in Asia, Africa and Latin America (Nguyen, et al. [20,21]). especially in children (Hunter [22]). Due to antimicrobial resistance of bacteria E. coli become a problem worldwide (Bayu Dharma S [23]). Among thousands of serotypes of E. coli species pathogenic E. coli harbor virulence factors, such as adhesins, invasins, entero- and cyto-toxins encoded by extra chromosomal plasmids, chromosomal pathogenicity islands, or bacteriophage united virulence factors for conquering host defences in order to cause intestinal and extra-intestinal diseases and known as diarrhoeagenic E. coli (Kaper, et al. [24]).
Multiplex Polymerase Chain Reaction (PCR) for Pathogenic Strain of E. coli
The results of multiplex PCR showed that, one isolate had band around 376 bp and another band was around 322 bp. Another study carried by (MUNSHI, et al. [25]). isolated diarrhoeagenic E. coli from surface water, among 21 isolates 5 isolates were randomly selected for PCR test and 3contained the shiga toxin (stx) gene while 2 to elt encoding heat-labile enterotoxin (LT), which indicate virulent gene encoding E. coli present in surface water. The only genes detected were eae and stx2, which were carried by 37.69% (n_248) of the isolates. Only eaewas harbored by 26.74% (n_176) of the isolates, representing potential a typical EPEC strains, while only stx2 was detected in 10.33% (n_68) of the isolates, indicating potential STEC strains (Chandran, et al. [14]).
Enterotoxigenic E. coli (ETEC) Isolates
Enterotoxigenic E. coli (ETEC) is a gram-negative diarrheal pathogen that infects almost 200 million people worldwide. (Gupta SK, et al. [26]). ETEC cause deaths almost 100,000 yearly and depend on a trio of ETEC virulence factors to cause disease (Hosangadi, et al. [27,11]). The prototypical ETEC strain H10407 contains heat-labile (LT) and heat-stable (ST) enterotoxins along with the colonization factor antigen I (CFA/I) adhesin (Evans, et al. [28,29]). dwelt on diarrheal epidemic caused by waterborne ETEC. From Bangladesh, one ETEC out of 27 E. coli, was detected in pond water (Apha, et al. [30,19]). observed that out of 144 E. coli isolates 10 were identified as toxigenic. In this study the virulent gene of ETEC were eltB and estA. And the isolated ETEC strain was collected in the summer season. In previous studies on the seasonality of ETEC prevalence, maximum isolation was in the warm and humid seasons and minimum in the winter months (Albert, et al. [8]).
Enterohemorrhagic and Enteropathogenic E. coli Isolates
In the present study, also identified another pathogenic strain of E. coli, which contained eaeA gene and gave band around 322 bp. This virulent gene can be present both in EHEC and ETEC strain. EHEC cause Haemorrhagic Urenic Syndrom (HUS) and EPEC infection causes urinary tract infection (UTI) which is primarily a disease of infants younger than 2 years (Nataro, et al. [4,5]). According to Clinical reports published by World Health Organization (WHO), a number of person infected with HUS die within 24 hours of symptoms displayed. Karmali, et al. [31]). reported the association of sporadic cases of hemolytic uremic syndrome with faecal cytotoxin and cytotoxin producing E. coli in stool. Among 40 E. coli isolates, one was eaeA gene, so the probability of getting this pathogenic strain was 2.5% (Pitcher, et al. [32]).
Pathogenic E. coli 0157:H7 is a deadly pathogen that is widely present in our country. The most common 0157: H7 outbreaks includes multistate outbreaks at 2006 and in Japan 1996 (Hadjilouka, et al. [29]). The higher recovery of E. coli indicates that this pathogen can survive outside their natural habitat for a long time. The biochemical properties of all strains of E. coli were not similar because of the genetic variation of this pathogen. The result of the MUG and CR-SMAC test indicates that all of their colony characteristics were not same also. The result of multiplex PCR indicates that pathogenic E. coli strain was also present. The virulent gene was eaeA, eltB and estA which represent the high incidence of diarrheal disease due to pathogenic E. coli. (Jalliffier-Verne, et al. [33]).
The authors express their gratitude to the Senior Scientist & Head of the Environmental Microbiology Laboratory, LSD, International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR, B). They are indebted for providing unrestricted support, essential to conduct the research. They are also thankful to all the members of the laboratory.


