Abstract
Since ancient times, medicinal plants have been widely utilized for their pharmacological properties. Their folklore uses for treatment of various ailments have been extensively studied in last decade. The Artemisia annua, Zingiber officinale and Hibiscus sabdariffa have long been studied for their antiviral effect. In this study, we conducted computational virtual screen of the active components of these plants against the SARS-CoV-2 to identify potential modulators. Active component of Hibiscus sabdariffa was found to possess good interaction energy with both the SARS-CoV-2 protease and spike protein. This study provides a standing ground to probe deeper into these plants for finding an herbal cure for the current SARS-CoV-2 outbreak.
Keywords: Computational Screen; Virtual Screen; Herbal Medicine; SARS-CoV-2; COVID-19
Abbreviations: WHO: World Health Organization; SARS-CoV: Severe Acute Respiratory Syndrome Coronavirus; CHL: Chloroquine; RMS: Root Mean Square; PDB: Protein Data Bank
Introduction
Viral diseases and their outbreaks posed threats to human
health and wellness and sometimes very deadly globally. Plants
have been found to be essential not only for food but also for
medicine, and the metabolism of plants is found to be the source
of many medicinal compounds for drug discovery. Many herbal
drugs have been used in clinics for hundreds or thousands of
years ones their safety and effects have been repeatedly tested
and can be immediately used during emergencies. Plant-derived
phytochemicals provide an excellent option for the safe treatment
of infectious diseases. Several recent articles [1-5] have emphasized
the importance of natural products from traditional medicine in the
drug discovery process and described their continued success in
contributing important molecules to the drug development pipeline.
Also, antiviral herbal medicines have been used in many historical
epidemics, and their analogs have been employed as the first line
of defense against viral diseases. In March 2020, the COVID-19
was declared a pandemic by the World Health Organization (WHO)
[6]. The virus beta coronavirus was identified to be related to
Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and
named SARS-CoV2 [7]. The outbreak was found to be more deadly
to people that have underlining chronic diseases linked to poor
nutrition. The antimalarial drug Chloroquine (CHL) and its safer
derivative Hydroxychloroquine (HCHL) have been proposed to be
a possible treatment for SARS coronavirus- 2 (SARS- CoV- 2) , the
causative agent of COVID- 19.
CHL /HCHL has anti-inflammatory activity and is used to treat
rheumatoid arthritis, lupus, and osteoarthritis. However, CHL
/HCHL has shown severe side effects, which may lead to heart
disease [8]. Accordingly, these developments have prompted us
to refocus our research on more safe and effective medications. Several kinds of essential oils and medicinal plants have been used
for many centuries for the treatment of respiratory infections,
asthma, dermatitis, and gastrointestinal diseases, and many studies
describe antiviral activities of plant essential oils (E.O.s) [9-17].
The multiple uses of E.O.s in the flock medicine encouraged many
investigators to isolate the possible active components and conduct
in vivo and in vitro studies on laboratory animals and human
beings, to understand its pharmacological action. These include
immune stimulation, anti-inflammatory, anticancer, antimicrobial,
anti-parasitic, antioxidant, antiviral, and hypoglycemic.
Materials and Methods
Ligands Preparation
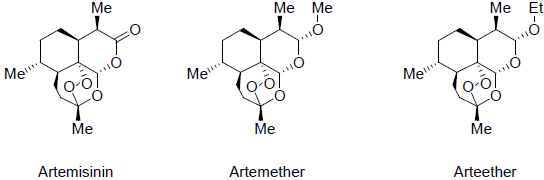
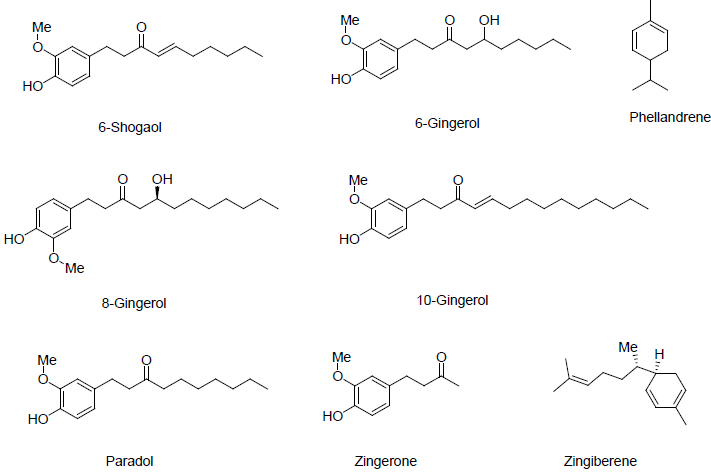
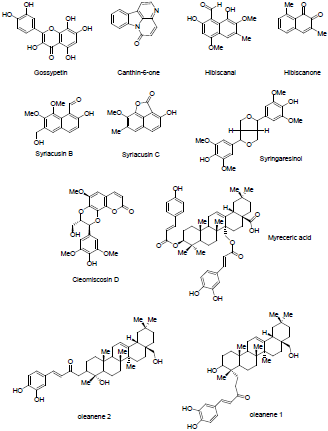
The structures of the active compounds isolated from these plant species were drawn into chem draw software [18] and exported to discovery studio (Figures 1-3). Ligands were prepared by adding hydrogens to all compounds, after which energy minimization was performed using the MMFF94x force field. Furthermore, ligand geometries were optimized using a Root Mean Square (RMS) gradient of 0.05 kcal/mol/Å using Discovery Studio 2020 [19].
Protein Preparation
The crystal structure of Sars-Cov-2 protease and spike protein were downloaded from the Protein Data Bank (PDB), using the PDB ID ‘6LU7’ for protease and ‘6M0J’ for spike protein respectively. The protein was then cleaned and extra chain with water molecules were removed. Minimization of the protein complex, to remove any constrain in the structure, was completed using prepare protein option in the software. Binding site was then generated, and ligands were docked using the C Docker [19].
Molecular Docking Studies to Illustrate the Importance of Plants in Treatment of Sars-Cov-2
To illustrate the importance of these plants, the active compounds isolated from these plants in the literature was used to study their possible effects in the current Sars-Cov-2 virus outbreak. Active compounds derived from these plants are illustrated in Figures 1-3. Molecular docking studies were implied to study the interaction of the most active compounds present in these three plant species and the important receptors of Sars-Cov-2 virus. Docking studies were performed using BIOVIA Discovery studio 2020 [19]. Crystal structure of Sars-Cov-2 protease and spike protein attached to the ACE2 receptor is available in Protein Data Bank (PDB) [20].
Validation of the Docking Results
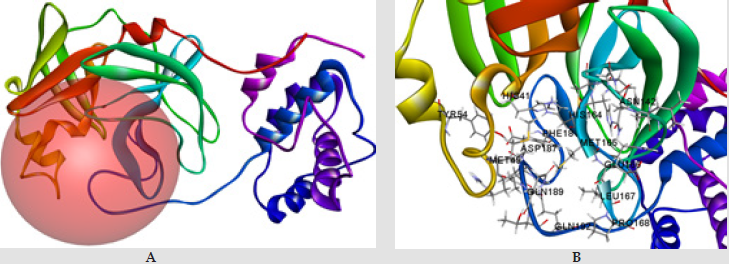
The docking experiments were validated by redocking of the co-crystal structure. The crystal structure of Sars-Cov-2 protease is present with the co-crystal structure. This co-crystal structure was used to build the binding site pocket. The co-crystal structure was then re-docked, to validate the results, with 10 posses. First five docking poses were selected and RMSD was calculate for each configuration. RMSD value was < 2 Å which is acceptable and indicate the validity of the prepared protein complex and docking protocol. The crystal structure of spike protein of Sars-Cov-2 is bound with ACE2 receptor (ACE2 receptor is used by the virus to enter the host cell). The ACE2 protein was removed, and spike protein was refined to build any missing loops. Binding site on the spike protein was then identified using the receptor cavities and the top binding site was selected for further docking (Figure 4). The prepared ligands were then docked in the prepared protein using the CDOCKER protocol with 10 poses of each ligand and pose cluster radius of 0.5 Å. CHARM m-based molecular dynamics algorithm is used by the CDOCKER tool to dock the ligand into the receptor protein and compute the interaction energies of the ligands. The interaction of the amino acids to the ligand was then viewed and saved in publication quality.
Figure 4: A; Spike protein and its active site determined by receptor cavity, B; interactive amino acids in the active site of spike protein.
Docking of Active Compounds in ACE2 Receptor Protein
Since the first outbreak of MERS in 2003 and the present Sars- Cov-2 outbreak in 2019, the coronavirus family have evolved and have become more dangerous and contagious. Another report published showed the evolution of current Sars-Cov-2 virus, suggesting a more severe second wave in the future. Most of the viruses belong to Coronavirus family use ACE2 receptor in the humans to enter the host cell. Although the evolution of virus protease and spike protein, these viruses use the same ACE2 receptor to gain host cell entry. ACE2 receptor being the target receptor to enter the host cell by the virus posit a viable target to stop the spread of Sars-Cov-2. Inhibiting ACE2 receptor will stop the viral entry into the host cell and replication. We used the crystal structure of ACE2 receptor protein bound with the Sars-Cov-2 spike protein (PDB ID 6M0J). The spike protein was removed and ACE2 receptor protein was refined to build any missing loops. Blind docking was then used to dock the most active compounds from the plant species to ACE2 receptor protein.
Results and Discussion
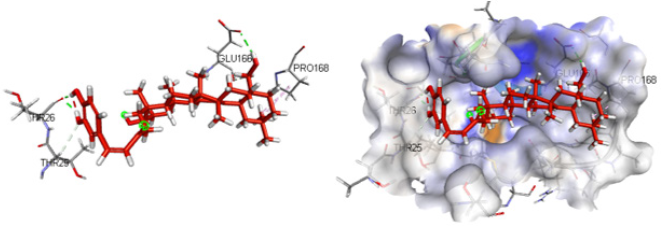
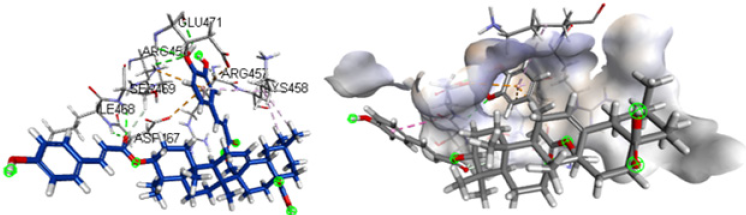
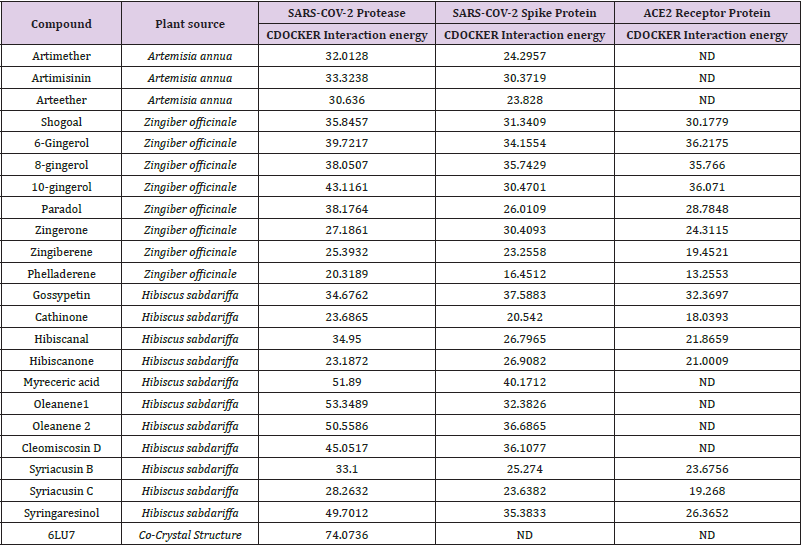
Table 1 describes the active compounds of the plant species with the CDOCKER interaction energies on the three different targets of Sars-Cov-2 virus. Higher values showed more interaction between the ligand and the amino acid of the target protein. The results showed that Oleanene 1 active compound of plant Hibiscus sabdariffa showed good interaction energy with protease enzyme of the coronavirus as compared to other active compounds of these plants. Oleanene 1 formed good interaction with amino acid THR25, THR26, PRO168 and GLY166 of the binding pocket. Similar hydrogen bond interaction with GLY166 and the co-crystal ligand was also seen. Figure 5 showed the compound Oleanene 1 fits perfectly within the binding pocket of Sars-Cov-2 protease binding site. Myreceric acid of the same species (Hibiscus sabdariffa) showed good interaction energy when docked in the Sars-Cov-2 spike protein. Inhibition of spike protein will halt the attachment of the virus into the host cell. Myreceric acid forms conventional hydrogen bond with amino acid ILE468, SER469, ARG454 and GLU471, Pi-bond with ASP467 and carbon-hydrogen bond with ARG457 amino acid (Figure 6). As discussed above, ACE2 receptor in the human is a viable target to stop the entry of virus into the host cell.
Figure 5: Compound Oleanene 1 (Hibiscus sabdariffa) in the active site of Sars-Cov-2 protease enzyme.
Figure 6: Compound Myreceric acid (Hibiscus sabdariffa) in the active site of Sars-Cov-2 spike protein.
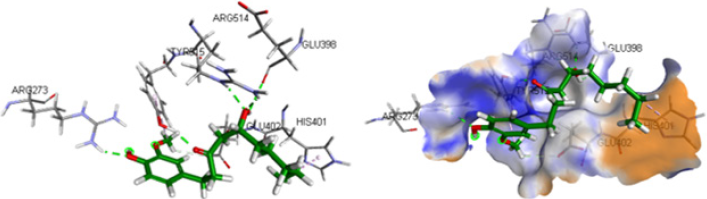
The docking results of the most active compounds of the three plant species with the ACE2 receptor protein showed that 6-Gingerol of plant Zingiber officinale possess good interaction energy with ACE2 receptor active site. Figure 7 showed the 6-gingerol in the active site of ACE2 receptor protein. 6-Gingerol showed hydrogen bond interaction with amino acid GLU398, ARG514, TYR515 and ARG273 of the active site. The docking results shows that Hibiscus sabdariffa plant is promising in killing the Sars-Cov-2 virus, as the active compounds of this plant showed good interaction energy with virus protease and spike protein. However, Zingiber officinale showed promising results to alter the entry of virus into the host cell by interacting with ACE2 receptor protein Table 1.
Conclusion
In conclusion, the study conducted showed that myreceric acid found in Hibiscus sabdariffa is effective in binding the active site of SARS-CoV-2 spike protein. Similarly, Oleanene1 extracted from the same species effectively binds to the SARS-CoV-2 protease. Since both the active compounds were found in Hibiscus sabdariffa, the use of this plant species to probe its effectiveness in the current SARS-CoV-2 outbreak should be further studied.
Acknowledgement
Authors would like to thank BIOVIA, Dassault Systems for providing free 6-month academic license of BIOVIA Discovery Studio, under “Working with Academics to End COVID-19” to conduct the computational studies. We are grateful to Robert A. Welch Foundation for support through departmental grant L - 0002.
Disclaimer
This study, in no shape and form, conclude that any plant extract can be used to treat SARS-CoV-2 virus. This provides a scientific starting point to look for the herbal alternative or active compounds for the treatment of virus.
References
- Makita C, Madala NE, Cukrowska E, Abdelgadir H, Chimuka L, et al. (2017) Variation in pharmacologically potent rutinoside-bearing flavonoids amongst twelve Moringa oleifera, Lam cultivars. S Afr J Bot 112: 270-274.
- Khalafalla MM, Abdellatef E, Dafalla HM (2010) Active principle from Moringa oleiferaLam leaves effective against two leukemias and a hepatocarcinoma. Afr J Biotechnol 9: 8467-8471.
- Mahmood KT, Mugal T, Haq IU (2010) Moringa oleifera: A natural gift-a review. J Pharm Sci Res 2(11): 775-2781.
- Vergara Jimenez M, Almatrafi MM, Fernandez ML (2017) Bioactive Components in Moringa oleiferaLeaves Protect against Chronic Disease. Antioxidants 6(4): 91.
- Shih MC, Chang CM, Kang SM, Tsai ML (2011) Effect of Different Parts (Leaf, Stem and Stalk) and Seasons (Summer and Winter) on the Chemical Compositions and Antioxidant Activity of Moringa oleifera. Int J Mol Sci 12(9): 6077-6088.
- (2020) "WHO Director-General's opening remarks at the media briefing on COVID-19". World Health Organization(WHO) (Press release).
- (2020) Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5: 536-544.
- (2020) Chloroquine paradox may cause more damage than help fight COVID-19; Sharma Anuj Microbes and infection (2020), under press.
- Schnitzler P (2019) Essential Oils for the Treatment of Herpes Simplex Virus Infections Quick View Other Sources By Schnitzler, Paul From Chemotherapy (Basel, Switzerland) 64(1): 1-7.
- Arino Shoko, Shirakami Hirohito, Hiruma Yukiko (2020) Virus inactivating agent containing essential oil and/or cationic starch, and external skin preparation composition Quick View. PCT Int Appl 21(10): 3426.
- Duschatzky C, Possetto M, Talarico L, García C, Michis C, et al. (2005) Evaluation of chemical and antiviral properties of essential oils from South American plants. Antiviral Chemistry & Chemotherapy 16(4): 247-251.
- Koch C, Reichling J, Schneele J, Schnitzler P (2008) Inhibitory effect of essential oils against herpes simplex virus type 2. Phytomedicine 15(1-2): 71-78.
- Saddi M, Sanna A, Cottiglia F, Chisu L, Casu L, et al. (2007) Antiherpevirus activity of Artemisia arborescens essential oil and inhibition of lateral diffusion in Vero cells 6: 10.
- Schnitzler P, Schuhmacher A, Astani A, Reichling J (2008) Melissa officinalis oil affects infectivity of enveloped herpes viruses. Phytomedicine 15(9): 734 -740.
- Wu QF, Wang W, Dai XY, Wang ZY, Shen ZH, et al. (2012) Chemical compositions and anti-influenza activities of essential oils from Mosla dianthera. Journal of Ethnopharmacology 139(2): 668-671.
- Garozzo A, Timpanaro R, Stivala A, Bisignano G, Castro A (2011) Activity of Melaleuca alternifolia (tea tree) oil on influenza virus A/PR/8: study on the mechanism of action. Antiviral Research 89(1): 83-88.
- Wu S, Patel KB, Booth LJ, Metcalf JP, Lin HK (2010) Protective essential oil attenuates influenza virus infection: an in vitro study in MDCK cells. BMC Complementary and Alternative Medicine 10: 69.
- Bicchi C, Rubiolo P, Ballero M, Sanna C, Matteodo M, et al. (2009) HIV-1-inhibiting activity of the essential oil of Ridolfia segetum and Oenanthe crocata. Planta Medica 75(12): 1331-1335.
- Mills N (2006) Chem Draw Ultra 10.0.
- BIOVIA, Dassault Systems, Discovery Studio v20, San Diego: Dassault Systems, 2020.

 Research Article
Research Article