Abstract
High resolution melting is a simple, efficient and cost-effective analysis, compared to alternative approaches used for SSR genotyping. However, it has not been evaluated yet, for its efficiency in high-throughput SSR genotyping, when large sample collections are to be analyzed. In such an approach, we tested its application to discriminate flavor-linked SSR markers, in a large collection of 211 grapevine varieties. Flavored and non-flavored varieties shared common alleles for the three markers VVMD27, VVC6 and VMC7G3, tested in this study. This is indicative of the redundant nature of the marker- linked QTLs along with other genetic determinants for expression of the flavor character in grapevine. The results showed that the three markers were highly polymorphic, detecting also SNPs in the sequences flanking SSR motifs. Such a high level of polymorphism was detected using the semi-automatic approach of HRM. Consequently, HRM proved to be powerful for distinguishing SSR genotypes, although repeated reference samples were needed for correct grouping, presenting a possible limitation of the method.
Keywords: Ηigh-Resolution Melting; Μicrosatellites; QTL; Vitis Vinifera L.
Abbreviations: SSRs: Simple Sequence Repeats; CE: Capillary Electrophoresis; HRM: High Resolution Melting
Introduction
Genetic diversity studies based on variation of simple sequence repeats (SSRs), have been greatly relied on easy to use gel-based techniques which, however, can be both time-consuming as well as inefficient for accurate genotyping [1]. Other techniques, like capillary electrophoresis (CE) requires post-PCR handling and is expensive. As an alternative, high resolution melting (HRM), a technique that monitors the decrease of fluorescence of a DNAbinding dye during the dissociation of double- stranded DNA [2], can provide an efficient solution for SSR genotyping at a reasonable cost. This measurement occurs immediately after PCR in the same tube at one-step. It is a rapid, simple, closed- tube, homogenous, and cost-efficient approach, that has high specificity and sensitivity, while allowing transition to high-throughput scale [3]. It has been widely applied in sequence polymorphism identification, as well as in the studies of genetic variability and genotyping [4-9]. However, its application, efficiency and limitations for high-throughput SSR genotyping have not been tested, yet.
Grapevine, Vitis vinefera L., is an important fruit crop worldwide grown for table grapes, wines, raisins and others. The presence of monoterpenes in its muscat varieties affects significantly the quality of their production [10,11]. Monoterpenes have been found both in grape leaves and berries in three forms: free volatiles, free polyhydroxylated molecules (polyols) and glycosidic derivatives of the two former types. Only the free volatile compounds make a direct contribution to the aroma, whereas free polyols and glycosidic derivatives constitute a reserve of odorless precursors, which generate flavor upon hydrolysis. Volatile monoterpenols, particularly, linalool, geraniol, nerol, citronellol and terpineol, have been found in much higher levels in the berries of the muscat varieties than the berries of the non-muscat ones (so-called neutral cultivars varieties) and exhibit a rather particular aroma related to this volatile composition [11,12]. Linalool, geraniol and nerol were found to be the three most abundant volatile monoterpenes [13], even though alone are not sufficient to generate the complete muscat aroma [14,12].
At the genetic level, muscat aroma in grapevine has been evaluated and a major QTL was identified on LG5, co-located with SSR markers including linked VVMD27 and VVC6 [11,12]. Dogligez [12], subsequently, mentioned a candidate gene within this major QTL discovered, encoding 1-deoxy-D- xylulose-5-phosphate synthase class 1 (DXS) for the content of volatile and non-volatile forms of geraniol, nerol and linalool [15]. It was demonstrated the co-localization of this aforementioned gene with the major QTL positioned on chromosome 5 considered to be the first enzyme in the plastidial methyl-erythritol-phosphate (MEP) pathway of terpene biosynthesis [18], in addition to its role in the monoterpenoid accumulation in the grape berry [11,16,17]. Another marker, VMC7G3 was identified to be localized to minor QTLs controlling the accumulation of linalool on LG 2 [11]. Furthermore, this marker was also proposed to be linked to the color locus [19,20]. Herein, is a first attempt to evaluate the capacity of the HRM technology for genotyping flavor linked SSRs in a large collection of grapevines, in order to verify its potential for high-throughput analysis of genetic variability and its efficiency in separating aromatic and non-aromatic grapevine varieties. Results of this study could also provide a useful marker methodology to assist early selection of grapevine offspring in breeding programs.
Materials and Methods
Plant Material
A total of 211 grapevine accessions (Table 1), single adult trees, were investigated in this study. The majority (200 varieties) derived from the Greek variety collection (including 4 cosmopolitan foreign varieties: Merlot, Cabernet Franc, Victoria, Muscat Hamburg) of the Hellenic Agriculture Organization - Demeter (HAO-Demeter); eleven derived from a Palestinian private vineyard. Leaf material was collected from actively growing parts of the trees and stored directly at −20°C until DNA extraction.
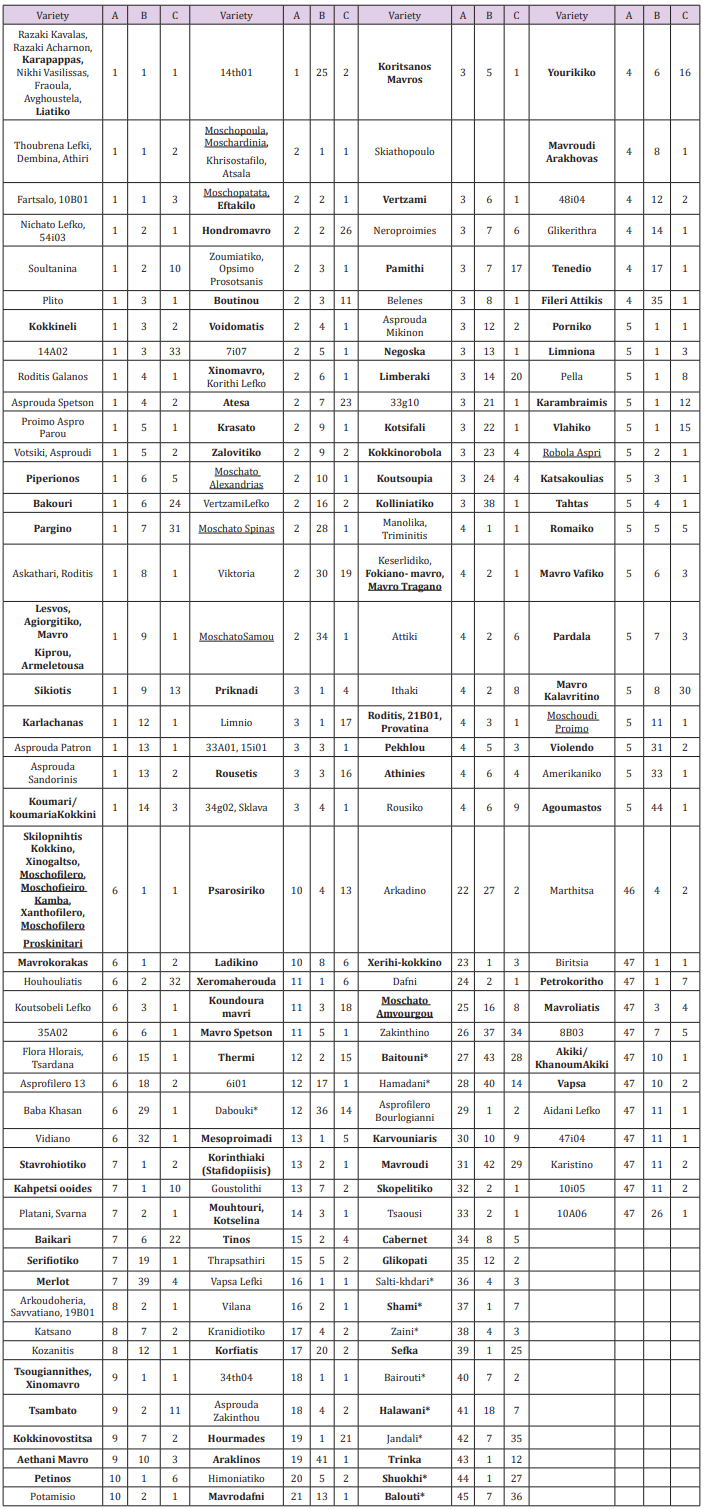
Table 1: List of Vitis vinifera genotypes analyzed. Each variety is presented by its common name while with a code are varieties from a breeding program; varieties underlined are known as having the muscat aroma; varieties bolded are colored; varieties ended with star are Paletinians. The HRM grouping for markers are A for VvMD27, B for VVC6, and C for VMDC7G3.
DNA Extraction and PCR-HRM
Genomic DNA was extracted following a modified CTAB protocol [21] that omitted the use of activated charcoal. All DNA samples were kept at 4o C for further use. The quantity and quality were then verified by a Nano Drop 1000 spectrophotometer (Thermo Scientific, USA) and 1.0% agarose gel electrophoresis, respectively. Three markers (VvMD27, VVC6 and VMC7G3) were used in this study [22]. PCR and HRM were performed using the LightCycler®96 Real-time PCR System (Roche Diagnosctics, Germany).Three 96- well plates were used for the analysis of the 211 samples for each marker. The final reaction volume was 11μL containing 1x KAPA HRM fast master mix (KAPA Biosystems), 0.25μM of each forward and reverse primer, 2.5 mM magnesium chloride, 10 ng of DNA template and filled up to 11μL with double-distilled water. The thermal cycling conditions involved enzyme activation at 95o C for 600 seconds, followed by 45 cycles of denaturation at 95o C for 30 seconds, annealing at 61o C for 30 seconds, and extending at 72o C for 40 seconds. Prior to the melting analysis, heteroduplex formation was enhanced by heating the amplification products at 95o C for 60 seconds and cooling down to 40o C for 60 seconds. For the HRM analysis, the plate was heated from 65o C to 97o C allowing 25 data acquisitions per 1o C.
HRM Data Analysis
HRM data analysis was performed using the LightCycler® 96 SW 1.1 software (Roche Diagnostics, Germany). All analyses were carried out including both positive (on the second and the third plates/SSR marker) and negative controls. The positive controls represented the different groups detected mostly on the first, then the second and then the third plates, and finally on a forth plate in case a new different group was discovered at the third plate. They were used to enable grouping the accessions among the three runs of each marker because the software does not support analyzing all data gathered from these runs together. After normalization, samples having the same melting curves were grouped automatically, with sensitivity settings at the default level 50%. However, grouping was also evaluated by visual inspection of the HRM profiles produced and corrected manually if needed through the adjustment of the pre and post-melting ranges by dragging the two leftmost and the two rightmost vertical sliders to appropriate locations. Generally, they were adjusted as close as possible to the melting temperature.
Sequencing and Mutation Discovery
PCR products were submitted for automated sequencing (CeMIA, Larissa, Greece). SNP discovery was conducted using Chromas Lite 2.1 (Technelysium Pty Ltd., Australia) and BioEdit software [23] to detect and characterize the polymorphisms among the sequencing results.
Results
All three markers amplified small amplicons less than 250bp. Furthermore, they produced polymorphic melting curves and revealed many different groups of genotypes. Both, visual inspections together with the classification produced by the LightCycler® 96 SW 1.1 software were used to perform the analyses in this study. VvMD27 marker was the most polymorphic, dividing the studied 211 varieties into 47 groups based on the number of different HRM profiles (Table 1). The largest group (1) comprised of 38 varieties none of which was aromatic. Group 2 consisted of 22 varieties 6 of which were aromatic. The rest of the aromatic varieties located into four more groups two of which were unique (having only one variety). VVC6 marker divided the varieties into 44 groups. Group 1, the biggest, had 50 varieties (Table 1) 5 of which were aromatic. Group 2 composed of 27 varieties three of which were aromatic. The rest of the aromatic varieties located into five more groups two of which were unique. The HRM profiles of VMC7G3 marker pointedly separated the studied varieties into 36 groups (Table 1). Group 1 had 106 varieties and almost all (12) aromatic belong to this group except for one (MoschatoAmvourgou) that was unique.
The combined semi-automatic (automatic classification followed by visual inspection and confirmation) analysis of HRM profiles was undertaken because of the inherent impact of sensitivity settings (defined by the user) for the automated algorithm on group calling. According to the instrument’s manual an increase of sensitivity subdivides the analyzed curves into more groups. Small Tm differences are valuated higher to separate curves into different groups. Also, small differences in curve morphology (for example, small shoulders) are valuated higher to separate curves into different groups. The instrument’s default sensitivity is set at 50% .Thus, sensitivity adjustments along with visual inspection of three different curves produced by the software (namely: Normalized Melting Curves, Difference Plot and Normalized Melting Peaks) lead to more conclusive discrimination of the different varieties. An almost perfect match in normalized melting curves and normalized melting peaks was considered as additional (to the software’s results) requirement for grouping varieties in the same group. In conclusion, results from automated classification needed correction for 33 genotypes, 13 of which were for marker VVMD27, 11 for marker VVC6 and 9 for marker VMC7G3.
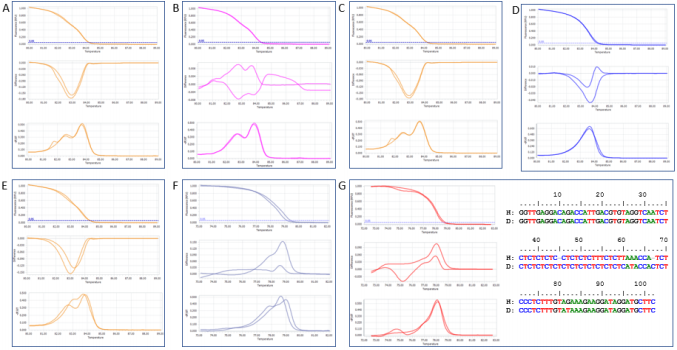
Following are several characteristic examples of group calling. The varieties Vertzami and Cabernet Franc have the same alleles (182bp and 190bp) [24] and produced similar but not identical HRM profiles of VvMD27 marker; they were automatically classified in the same group (Figure 1A). Kotsifali and Limnio carry the same alleles (186bp and 190bp) [24] and were identical in their HRM melting curve and melting peak profiles although different at the shape of the difference plot (difference plot curves may be quite different in samples classified in the same group, depending on the baseline samples used for reference curve subtraction); they were automatically placed in the same group (Figure 1B). Other varieties, for example Merlot and Cabernet Franc (Figure 1C), Vilana and Soultanina (Figure 1D), Goustolithi and Vertzami (Figure 1E) have different alleles [24] and showed differences in their HRM profiles. However, automatic group calling classified these varieties in the same group. These were classified in different groups when performed the combined analysis.
In addition, Balouti* and Shami* (Figure 1F) and Dabouki *and Halawani* (Figure 1G) are other example pairs with obvious differences in their HRM profiles, but mis-autocalling in the same group of VMC7G3 and VVC6 markers, respectively. To validate the results of the combined analysis of HRM profiles, representative samples of some genotype pairs were sequenced. As shown in Figure 1G, sequence alignment of the PCR products from Dabouki* and Halawani* with the marker VVC6 depicted the differences among these genotypes as well as the incorrect automatic classification within the group that was adjusted by visual inspection of the curve shapes. Highlighting the Palestinian varieties, none of which considered to be aromatic, they were randomly distributed among the Greek ones in case of both VVC6 and VMC7G3 analyses; however, they were particularly unique in VVMD27 analysis.
Figure 1: Comparison of HRM profiles (Top: Normalized Melting Curves, Middle: Difference Plot and Bottom: Normalized Melting Peaks) of different variety/marker combinations with characteristic results for variety grouping. Profiles of A: Vertzami and Cabernet Franc, B: Kotsifali and Limnio, C: Cabernet Franc and Merlot D: Vilana and Soultanina E: Goustolithi and Vertzami for markerVVMD27; F: Balouti* and Shami* for VMC7G3; G: Dabouki* and Halawani* for VVC6 along with sequence alignment.
Discussion
HRM has been proven an efficient and inexpensive approach to detect sequence variations in humans, plants and microorganisms at the SNPs level. In addition, it requires less technical expertise and hands-on time and excludes hazardous substances encountered in capillary electrophoresis or other detection technologies [25]. Recently, HRM technology has been exploited to detect SSR variation in plants. It was used for genotype identification and authenticity testing [5,6,26,27]. Moreover, it can also be used for mutation scanning [29]. Consequently, such a technology has been considered as an effective approach to genotype using SSR markers at the inter-specific level because similarity is still very high among such plants [5]. However, high-throughput HRM genotyping of sample numbers exceeding the capacity of a single multi-well plate has not been reported.
In the present study, we applied a semi-automated analysis of HRM data which mostly facilitated the conclusive discrimination among a large number of varieties exploiting the sensitivity of normalized melting curve, normalized melting peak and difference plot profiles. Distefano et al., 2012 also suggested such a technique if the population under investigation is large and involves many species [5]. Our results demonstrated the efficiency of this approach and uncovered its strengths and limitations. HRM proved to be a powerful method for distinguishing SSR genotypes and uncovering SNPs in the sequences flanking SSR repeat motifs.
However, analysis of a large collection of samples necessitated positive controls for each group of genotypes detected among the different runs of each marker. In this study we used the LightCycler® 96 (Roche, Germany), a medium cost instrument with 96-well plate capacity. To exceed this capacity one must look for a much more expensive 384-well instrument (such as LightCycler 480), although the sample limit is still dependent on its Peltier-block capacity. Thus, the need for appropriate positive controls is a significant limitation for high-throughput SSR genotyping if large collection of samples must be analyzed. Another limitation encountered in this study was the requirement of sequencing, contrary to the principle of the approach, when inconclusive HRM profiles were encountered. This was due to the sequence complexity of microsatellites when high number of alleles are present in the analyzed material. Such complications have also been reported in nearly all the studies carried out using HRM to analyze SSR variation in plants, even though they were performed for smaller number of samples [4-9]. HRM has been supposed to be more effective for low complexity SSRs with low number of alleles to ensure reliable interpretations of the melting curve profiles [6].
Regarding the aroma-linked markers, the HRM analyses of both VVMD27 and VVC6 markers derived rather similar results, grouping the analyzed varieties into large number of genotypes. In addition, most of the flavored varieties grouped together with nonflavored ones, although some of them were unique. Incorporation among flavored and non-flavored varieties was due to presence of the same alleles as confirmed by sequencing (data not shown). This is indicative of the redundant nature of the VVMD27 and VVC6 markers along with other genetic determinants for expression of the flavor character in grapevine. VVC6 separated the aromatic varieties into 7 groups while VVMD27 gave rise to 5 groups. Such a result could be due to their locations, the VVC6 being closer to the QTL (VVC6-Ch5:441633-441654; VVMD27- Ch5:4472022- 4472200; aroma QTL-Ch5: 3851143-3856236). However, the distance between them might explain the grouping detected among the aromatic and non-aromatic varieties based on small quantities of monoterpenes which are under the odor detection threshold in the non-aromatic ones [30].
Recently, analysis of a collection of 122 Greek varieties, many of which were common with this study, classified the muscat varieties according an SNP polymorphism of the VvDXS gene coding region regarding an SNP 1822G>T substitution, grouping correctly 15 muscat varieties and identifying the substitution in 2 non-muscat varieties [31]. Accordingly, Wagner [28] mentioned that the genetic control of muscat flavor was associated with at least five complementary dominant genes plus a modifier gene. In addition, these results might be due to pleiotropic effects of genes affecting the content of different aromatic compounds among the aromatic and non-aromatic varieties. These regulatory loci may occur in different genetic backgrounds [11]. VMC7G3 divided the varieties into a smaller number of different groups compared to the other markers. However, 12 out of 13 aromatic varieties were found in one group together with other non-aromatic ones, suggesting the origin of the allele from a non-aromatic ancestor. According to Doligez et al. [12], the origin of variability for the linalool QTL on LG 2 in a mapping population was the non-muscat parent. Then, the linalool content in this parent is under the odor detection threshold or the favorable allele from this parent is recessive. Furthermore, similarities detected among the muscat varieties here could be explained by the narrow genetic basis of the Muscat family [32]. Contrary to Doligez et al. [12] findings, Battilana et al. [11] found no genes underlying the QTL for linalool content on LG 2. However, they proposed the presence of genes acting downstream in the biosynthetic pathway of linalool, such as the (S)-linalool synthase or other regulatory loci.
Conclusion
In conclusion, the present study represents the first attempt to use HRM profiling in order to analyze a large collection of genotypes with SSR markers. Strengths and caveats of the approach are indicated and a semi- automatic approach that may be adapted for the used instrument is proposed. Our results demonstrate that HRM-PCR could be a better method than gel-based SSR genotyping and also more cost-effective, simple and efficient from CE. It has the disadvantage that it needs reference samples for all groups that are identified in a 96-well plate when the collection is larger and multiple runs for the same marker are required in a highthroughput format of the assay. However, it is simple, efficient and cost-effective for a moderate number of samples that are usually examined in plant genotyping research.
References
- Yang M, Yue YJ, Guo TT, Han JL, Liu JB, et al. (2014) Limitation of highresolution melting curve analysis for genotyping simple sequence repeats in sheep. Genetics and Molecular Research 13(2): 2645-2653.
- Reed GH, Kent JO, Wittwer CT (2007) High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 8(6): 597-608.
- Vossen RH, Aten E, Roos A, Den Dunnen JT (2009) High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat 30(6): 860-866.
- Arthofer W, Steiner FM, Schlick Steiner BC (2011) Rapid and costeffective screening of newly identified micro satellite loci by high resolution melting analysis. Mol Genet Genomics 286(3-4): 225-235.
- Distefano G, Caruso M, La Malfa S, Gentile A, et al. (2012) High resolution melting analysis is a more sensitive and effective alternative to gelbased platforms in analysis of SSR - an example in citrus. PLoS One 7(8): e44202.
- Ganopoulos I, Argiriou A, Tsaftaris A (2011) Microsatellite high resolution melting (SSR- HRM) analysis for authenticity testing of protected designation of origin (PDO) sweet cherry products. Food Control 22(3-4): 532-541.
- Mackay JF, Wright CD, Bonfiglioli RG (2008) A new approach to varietal identification in plants by microsatellite high resolution melting analysis: application to the verification of grapevine and olive cultivars. Plant Methods 4: 8.
- Mader E, Lukas B, Novak J (2008) A strategy to setup codominant microsatellite analysis for high-resolution-melting-curve-analysis (HRM). BMC Genet 9: 69.
- Muleo R, Colao MC, Miano D, Cirilli M, Intrieri MC, et al. (2009) Mutation scanning and genotyping by high-resolution DNA melting analysis in olive germplasm. Genome 52(3): 252-260.
- Ribéreau Gayon P, Glories Y, Maujean A, Dubourdieu D (2000) The chemistry of wine: stabilization and treatments. In: Wiley J and Sons Ltd (eds.). Handbook of enology 2: 187-206.
- Battilana J, Costantini L, Emanuelli F, Sevini F, Segala C, et al. (2009) The 1-deoxy-D-xylulose 5-phosphate synthase gene co-localizes with a major QTL affecting monoterpene content in grapevine. Theor Appl Genet 118(4): 653-669.
- Doligez A, Audiot E, Baumes R, This P (2006) QTLs for muscat flavor and monoterpenic odorant content in grapevine (Vitis vinifera L.). Mol Breeding 18(2): 109-125.
- Ribérau Gayon P, Boidron JN, Terrier A (1975) Aroma of muscat grape varieties. J Agric Food Chem 23: 1042-1047.
- Bayonove C, Cordonnier R (1971) Recherches sur l’aroˆ me du muscat. III. Etude de la fraction terpenique. Ann Technol Agric 20: 347-355.
- Cordoba E, Salmi M, León P (2009) Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J Exp Bot 60(10): 2933-2943.
- Duchene E, Butterlin G, Claudel P, Dumas V, Jaegli N, Merdinoglu D (2009) A grapevine (Vitis vinifera L.) deoxy-D-xylulose synthase gene colocates with a major quantitative trait loci for terpenol content. Theor Appl Genet 118(3): 541-552.
- Lois LM, Rodríguez Concepción M, Gallego F, Campos N, Boronat A (2000) Carotenoid biosynthesis during tomato fruit development: regulatory role of 1-deoxy-D-xylulose 5- phosphate synthase. Plant J 22(6): 503-513.
- Emanuelli F, Battilana J, Costantini L, Le Cunff L, Boursiquot JM, et al. (2010) A candidate gene association study on muscat flavor in grapevine (Vitis vinifera L.). BMC Plant Biology 10: 241.
- Lijavetzky D, Ruiz Garcia L, Cabezas JA, De Andres MT, Bravo G, et al. (2006) Molecular genetics of berry colour variation in table grape. Mol Gen Genom 276(5): 427-435.
- Pelsy F, Dumas V, Bevilacqua L, Hocquigny S, Merdinoglu D (2015) Chromosome Replacement and Deletion Lead to Clonal Polymorphism of Berry Color in Grapevine. PLOS Genetics 11(4): e1005081.
- Krizman M, Jakse J, Baricevic D, Javornik B, Prosek M (2006) Robust CTAB-activated charcoal protocol for plant DNA extraction. Acta Agric Slovenica 87(2): 427-433.
- https://www.ncbi.nlm.nih.gov/probe
- Hall TA (1999) Bio Edit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95-98.
- http://www.vivc.de/
- Li XH, Acharya A, Farmer AD, Crow JA, et al. (2012) Prevalence of single nucleotide polymorphism among 27 diverse alfalfa genotypes as assessed by transcriptome sequencing. BMC Genomics 13: 568.
- Jaakola L, Suokas M, Häggman H (2010) Novel approaches based on DNA barcoding and high-resolution melting of amplicons for authenticity analyses of berry species. Food Chem 123(2): 494-500.
- Madesis P, Ganopoulos I, Argiriou A, Tsaftaris A (2012) The application of Bar-HRM (Barcode DNA-High Resolution Melting) analysis for authenticity testing and quantitative detection of bean crops (Leguminosae) without prior DNA purification. Food Control 25(2): 576- 582.
- Wagner R (1967) Etude de quelquesdisjonctions dans des descendances de Chasselas, Muscat Ottonel et Muscat à petits grains. Vitis 6: 353-363.
- Vorkas PA, Christopoulos K, Kroupis C and Lianidou ES (2010) Mutation scanning of exon 20 of the BRCA1 gene by high-resolution melting curve analysis. Clin Biochem 43(1-2): 178-185.
- Baek HH, Cadwallader KR (1999) Contribution of Free and Glycosidically Bound Volatile Compounds to the Aroma of Muscadine Grape Juice. Journal of Food Science 64(3): 441-444.
- Merkouropoulos G, Ganopoulos I, Doulis A, Nikolaou N, Mylona P (2016) High Resolution Melting (HRM) analysis on VviDXS to reveal muscats or non-muscats among autochthonous Greek wine producing grape varieties. OENO One 50(3): 161-167.
- Crespan M, Milani N (2001) The Muscats: a molecular analysis of synonyms, homonyms and genetic relationships within a large family of grapevine cultivars. Vitis 40(1): 23-30.

 Research Article
Research Article