Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Revati Deshmukh1 and Priya Nimish Deo2*
Received: January 16, 2018; Published: January 22, 2018
*Corresponding author: Priya Nimish Deo, Asst Professor, Department of Oral Pathology and Microbiology, Bharati Vidyapeeth Deemed to be University, Dental College and Hospital, Pune, Maharashtra, India
DOI: 10.26717/BJSTR.2018.02.000679
Proteomics and Genomics have opened a new era in the field of research and technology. This has made possible molecular profiling of normal and diseased states. Identification of specific proteins in a particular disease will provide information for drug development which will interfere with the action of those proteins. This will help in establishing targeted therapies. Personalized medicine will provide treatment based on the genetic makeup of an individual and by taking into account the variations noted in a patient. Translation of this research in diagnostics and treatment planning is essential for the benefit of the patient.
Abbreviations: HNSCC: Head and Neck Squamous Cell Carcinoma; LOH: Loss of Heterozygosity ; CYP: Cytochrome P450; GSTM: Glutathione -S-Transferase Genes; UGT: UDP-glucuronosyl transferase; MMP: Matrix Metalloproteinases; Ig: Immunoglobulins; TNF: Tumor Necrosis Factor; CHS: Chediak- Higashi Syndrome; SNP: Single Nucleotide Polymorphism ; 2D PAGE: 2- Dimensional Polyacrylamide Gel Electrophoresis
With the upsurge of personalizing virtually anything such as mugs, stationery, T-shirts, phone cases, gift items it is not surprising that medicine is fully taking root in this domain. Over the years, there has been a gradual paradigm shift from traditional medicine as a result of increase in scientific knowledge. The traditional path of drug development which has conventionally influenced the practise of medicine has been based on identifying therapies which target an entire population [1]. With advances in research and technology it will be possible for health care professionals to treat patients based on their genetic constitution - a practise called as personalized medicine. Genomics and Proteomics have promised to change the practice of dentistry and Oral Pathology, allowing the identification and characterization of risk factors and therapeutic targets at a molecular level. However, if compared to other areas of medicine, the progress in oral pathology achieved using purely genomic approaches have been overall limited. It has been suggested that other “-omics” disciplines, such as proteomics and metabolomics, should be applied to reach a deeper understanding of the molecular mechanisms underlying oral disorders [2].
Genome is the entire genetic makeup of the human cell nucleus or the sum total of all an individual organisms’ gene. Genome is defined as the master blue print for cellular structures and activities during the life time of each and every cell; the genome contains the complete set of instructions for the initiation, construction, operation, maintenance and repair of all living organisms. Human genomics is the study of structure, functions and interactions of all genes in the human genome that promises to improve the diagnosis, management and prevention of disease [3]. While recent estimates suggest there are approximately 50,000 genes, there are thought to be three times that many proteins as a result of alternative splicing and post-translational modification [4]. In addition to the genomic information found within nucleus of each human reproductive cell (sperm in testes and ova in ovaries) as well as the trillions of somatic cells. (eg – cartilage, bone, PDL, dental pulp, trigeminal ganglia, salivary glands, oral mucosa), genomic information is also encoded within genes located in the maternally inherited mitochondria termed the mitochondrial genome or mit DNA Figure 1.
With the exception of trauma, essentially all diseases and disorders have a major genetic component. Human diseases and disorders may result from single gene mutations, but more commonly result from complex and multiple gene-genes and geneenvironment interactions [5]. The term proteome indicates the total set of proteins encoded by a genome. Proteomics involves the surveying of global protein composition of a cell or organism. It is necessary to monitor the level along with the activity of proteins. This field incorporates technologies that can be applied to serum and tissue in order to extract important biological information in the form of biomarkers to aid clinicians and scientists in understanding the dynamic biology of their system of interest, such as a patient with cancer. Current goals of proteomic research are more varied and directed towards the systematic determination of diverse properties of proteins in various physiological and pathological conditions [6].
Figure 1: Review of technologies used for identification of proteins [9].
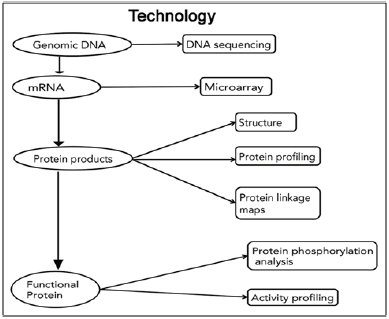
Many oral diseases and disorders, including dental caries, cleft lip/palate, and a host of craniofacial syndromes are complex conditions that arise from the actions of multiple genes and their interactions with one another, the environment and other factors. It will take more than adding up the molecular parts to understand and address such multi-factorial disorders. An interdisciplinary approach is needed to integrate the complex web of molecular information with clinical information, particularly for diseases where diagnosis is based primarily on clinical findings. Management of head and neck cancer is still based primarily on the evaluation of macroscopic tumor characteristics and extent of disease. But if genomic information could be factored into treatment decisions, it might be possible to predict which patients require aggressive treatment and which do not. Surgeons might be able to distinguish healthy tissue more precisely thus minimizing disfiguration and loss of function [7].
Oral Squamous Cell Carcinoma is the sixth most common malignancy worldwide. Squamous cell carcinoma appears to reflect sequential and multiple mutations within complex pathways overtime in response to multiple gene environment interactions often associated with direct sunlight, alcohol consumption, chronic use of tobacco products, chronic infection and immune and nutritional deficiencies. The biological problem of squamous cell carcinoma found in oral cancer patients likely demonstrates multiple mutations in discrete motifs that directly regulate cell division, cell adhesion, cell communication and/or programmed cell death or apoptosis. These new techniques provide an approach to unravel these possibilities and to focus upon candidate gene products for gene therapy or gene mediated therapeutics [5].
In contrast to germ line mutations that cause genetic syndromes, head and neck squamous cell carcinoma (HNSCC) are characterised by genetic alterations in somatic cells such as loss of heterozygosity (LOH), mutations, amplification (and hence overexpression) of certain genetic regions and genomic re-arrangements. These genetic changes appear to be early and essential events in tumour development. Genetic alterations consistently correlated with histological stages of head and neck squamous cell carcinoma have been used to develop molecular progression models for the disease. Genetic polymorphisms of several xenobiotic metabolizing enzymes including the cytochrome P450 1A1 (CYP1A1), glutathione -S-transferase genes (GSTM1 and GSTT1) and UDP-glucuronosyl transferase 1A7 (UGT1A7) genes may confer increased risk for tobacco related HNSCC.
Understanding the spectrum of genetic susceptibility for HNSCC may help identify those agents that increase risk for disease and permit identification of those individuals at greatest risk. For example, alterations of the tumour suppressor gene p53 are reported in a variety of cancer types. This physiologically important gene may also be affected by human papilloma viruses, which appear to have a causal association with a subset of head and neck cancers. Altered expression of several of the matrix metalloproteinases and their inhibitors may be important determinants of the invasiveness and ability to metastasize seen in HNSCC. It is likely that genetic characterization will be combined with more traditional histological staging of HNSCCs to develop classifications of better clinical utility [4].
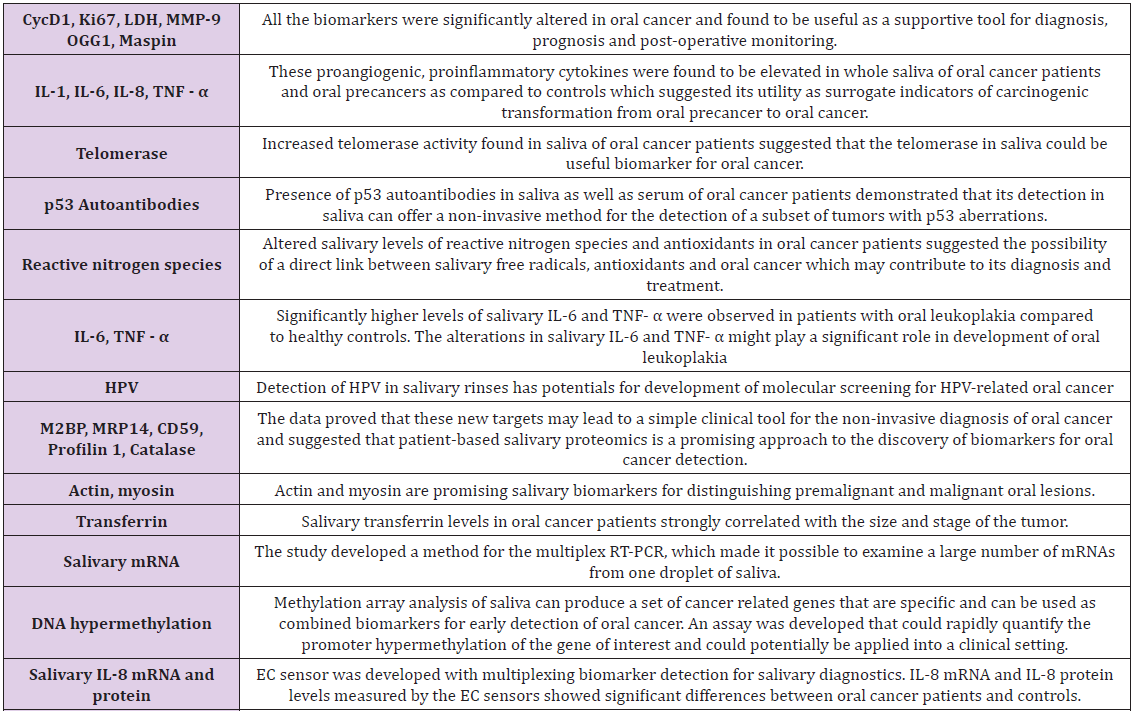
The high morbidity and mortality rate in OSCC patients ultimately led to the aspiration for enhanced knowledge of the characteristics and pathogenesis involved, as clinical and histological analysis are the only basis for OSCC diagnosis, furthermore, specific biomarkers are highly supportive of untraceable hidden sites of OSCC and for screening of high risk patients. In comparison to tissue biopsy which is invasive, there have been plenty of biomarkers identified, amongst them, the most recommended biomarker detection medium for OSCC includes biomarkers in body fluids, such as blood and saliva (Table 1) [8,9].
Table 1: Biomarkers of saliva for cancer diagnosis [9].
Periodontal ligament fibroblast protein expression has been studied using immunological methods, although this technique is limited to previously identified proteins for which specific antibodies are available. A total of 117 proteins have been identified from PDL fibroblasts which can serve as a reference map for future clinical studies as well as basic research. Periodontal diseases are still worldwide human ailments, resulting in a high level of morbidity and an economic burden to the society. Proteomics offers a new approach to the understanding of holistic changes occurring as oral micro-organisms adapt to environmental changes within their habitats in the mouth. Porphyromonas gingivalis is a periodontal pathogen, is known to undergo a transition from its commensal status in healthy individuals to a highly invasive intracellular pathogens in human patients suffering from periodontal diseases. Extensive proteomic research is done on P. Gingivalis [10].
Saliva is considered as an important periodontal diagnostic tool since variable amounts of blood, serum, serum products, GCF, electrolytes, epithelial and immune cells, micro-organisms, bacterial degradation products, lipopolysaccharides, bronchial products and other foreign substances are present in whole saliva. Matrix metalloproteinases (MMP2, 39), Immunoglobulins (Ig), esterases, lysozyme, lactoferrin levels in saliva are valuable for predicting the progression of periodontitis. Various cytokines like C-reactive proteins, pentraxin-3, TNF, various other interleukins which are involved in its pathogenesis have come handy in diagnosing periodontal diseases [10].
A direct cause and effect relationship for a specific gene defect and periodontitis susceptibility can be illustrated by the association of severe periodontitis with a number of genetic diseases. Over the past decade, the genetic basis for several syndromic forms of periodontitis has been identified. Mutations of the cathepsin C gene are responsible for Papillon-Leferve syndrome, Haim Munk syndrome, and atleast a portion of pre-pubertal periodontitis cases. CHS gene mutations cause Chediak- Higashi syndrome and beta-2- integrin (a cell surface receptor) gene mutations are responsible for leucocyte adhesion deficiency type 1. While specific genes have been identified for several syndromic forms of periodontitis, the genetic basis for the more prevalent (-1-2 percent ) forms of aggressive periodontitis suggest that susceptibility is inherited as a simple genetic trait, but it is unclear how many genes may be involved in these non-syndromic forms of periodontitis.
While the major genes responsible for aggressive periodontitis remain elusive, several genes that may modify the clinical expression of this disease form have been identified including interleukin-4 polymorphism, Fc gamma receptor genotype, Vitamin D receptor polymorphisms and immunoglobulin allotypes. In addition to a significant genetic etiology for aggressive periodontitis, there is substantial evidence that environmental factors such as smoking and microbial agents (virulence factors) interact with modifying genes to determine final disease trajectory. In contrast to the relatively simple genetic etiology of syndromic forms of periodontitis, it is clear that aggressive periodontitis has a more complex etiology, and the final disease phenotype is modified by multiple genetic and environmental factors [4].
Dental caries is an infectious disease where there are numerous host resistance and risk factors that are genetically determined. Dental caries is influenced by numerous genomic factors, a mutation in SNP of Amel X, a gene coding for a protein, which is crucial for normal enamel development is correlated with increased caries susceptibility. Decrease in proline rich protein levels increases caries susceptibility of dental caries. Defect in KLK4 gene coding for a protein expressed during enamel maturation causes a decrease in the hardness of enamel thereby increases the susceptibility of an individual to caries. Identification of specific gene changes and interactions aids the dentist in identifying the caries risk individuals so that more frequent oral hygiene maintenance and aggressive treatment plan can be established [3].
Various technologies have been developed to identify proteomic measurement. Proteomic data are tremendously useful in classifying cells and tissues in disease states and understanding different biological mechanisms. Scientists identify the structure, interactions and functions of all proteins within the cells and organisms by utilizing methods of the protein measurement. The current focus in proteomics is to define the nature of proteins and their level of expression in cells using the protein detection technologies. Clinical samples of proteins can be analysed through procedures such as mass spectrometry, 2- Dimensional polyacrylamide gel electrophoresis (2D PAGE) and protein arrays in order to help compare the protein variability between samples [6].
Personalized medicine will provide treatment based on the genetic makeup of an individual by taking into account the genetic variations which are noted in a particular disease for that particular patient rather than standardized treatment for every patient. Translation of this research in a clinical set up will make it beneficial for the patient as well as the clinician in early diagnosis and treatment planning. It is advisable that if health care professionals are exposed earlier to research and technology, the clinical implications of various diseases will be channelized better, thus making application of research into clinical practise easier.


