Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Jean-claude Perez*
Received: July 17, 2017; Published: July 20, 2017
Corresponding author: DR Jean-claude Perez, Maths and Computer Science Retired interdisciplinary researcher (IBM), 7 avenue de terre-rouge F33127 Martignas, Bordeaux metropole, France
DOI: 10.26717/BJSTR.2017.01.000209
Keywords: Genomes Invariant; Periods; Genomes Unification
Starting from the fundamental basic level of Bio-Atoms atomic weights, we discovered a numeric code common to the 3 Genetics worlds: DNA, RNA and amino acids. Coding Genomics (DNA double strands) and “Virtual Proteomics”(corresponding amino acids translations of DNA), we obtain specific couples of curves called Master Code signatures; in all cases both curves are highly correlated, meanwhile, there are hierarchies between these Genomics/Proteomics coupling ratios: Analyzing and comparing Master Codes in Achaea’s, Bacteria’s, Viruses, and Eukaryotes reveals function (actives sites, SNPs locations etc), genomes integrity, differentiation and evolution new information [1-5]. We have data suggesting the Electrical nature of this Law. Specifically in the case of Human Genome, we prove triple quasi UNIFICATION between:
A. The 2 Genomics and virtual Proteomics bio-chemical materializations.
B. The 3 possible codons reading frames.
C. The 4 combinatorial possibilities readings directions of both DNA strands.
In fact, we demonstrate the evidence of a super UNIFIED information level including both Genomics and Proteomics worlds. We thus analyzed the 285 and the 66 millions base-pairs of human chromosomes 1 and 20 (more than 10% of the whole draft human genome). By analysing both chromosomes in independent units of 1Mbps, there are (285+66)x2x3x4 independent sequences which were studied (nearly 8.4 billion base-pairs). Then, there appears a master code structuring the complete draft human chromosomes 1 and 20 with an average Genomics/Proteomics coupling ratio of 97%. Then another level of reading reveals embedded long distance periodic structures throughout whole genomes [6-12]. These PERIODS, expressed in Codons numbers multiples, characterize globally specific Genomes: there are independent with classical nucleotides repeats. A possible meaning of this discovery could be related with DNA meta-structure at physical and spatial levels (DNA molecule on-rolling up). These discoveries constitutes a “shadedlanguage” called the “universal genomic code”. This fundamental discovery was first published in the book (4) then summarized by the main article (9), and was generalized to all genomes available today: archaeas, bacteria’s,viruses, worm, fly, Arabidopsis and the complete draft humangenome.
Figure 1: The 99.30% Genomics/Proteomics perfect coupling % of HIV1 CPZGAB isolate.
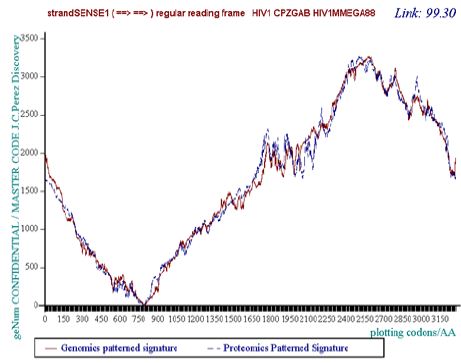
Like other viruses genomes, the HIV/SIV genomes provide Master Code coupling and periods. Meanwhile, the coupling is worse than in the case of the human genome. We have analyzed the master code of the 160 HIV1/HIV2/SIV genomes available from the LOS ALAMOS AIDS database. Our innovative results provide for each specific AIDS genome isolate:
A. MASTER CODE Genomics/Proteomics coupling signature (see (Figure1)- HIV1 CPZGAB isolate).
B. EVIDENCE of META-PERIODS overlapping whole genomes (see (Figure 2)- HIV1 NDK isolate).
Figure 2: The perfect 4-Codons META-PERIOD of HIV1 NDK isolate.
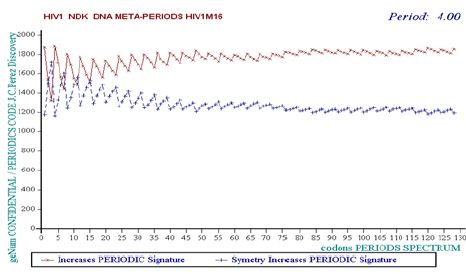
C. MASTER CODE Genomics/Proteomics average coupling for the better codons reading frame of the 160 complete genomes: 92.2%.
Average results for the 89 HIV1 isolates: 92.05% / Average results for the 28 HIV2 isolates: 91.31% / Average results for the 43 SIV isolates: 93.09% [13-15]. EVIDENCE of a common 4-codons META-PERIOD (12 nucleotides) Invariant Unifying all the 160 HIV/SIV referenced Genomes Isolates. Evidence of small PERIOD-4 variations in some specific isolates linked with possible heterogeneity/differentiation ways related to these Genomes [16].
The discovery of an invariant of very high level which either common to the totality of the 160 known HIV1, HIV2 and SIV variants Genomes constitutes a new fact. The study of the fluctuations of certain genomes around this PERIOD-4 reference could make it possible to build universal vaccines or therapies. The fact that this PERIOD-4 is specific to all the HIV retroviruses, differentiating them with other viruses, bacteria’s or human genomes, could be used towards an universal targeted AIDS vaccine.


